This website contains links to codes developed by Dr. Vannucci and her students and collaborators, together with some of the datasets used in their publications.
Codes for recent papers can be found at Dr. vannucci’s Github repository
Codes for some of Dr. Vannucci’s work on epilepsy research, with Sharon Chiang, Emily Wang and Robert Moss, can be found at the dedicated GitHub site Seizurerisk
Featured software:
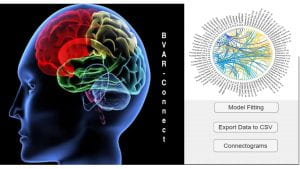
BVAR-Connect MATLAB GUI: User-friendly MATLAB GUI for Bayesian vector autoregressive models for effective connectivity network estimation based on resting-state fMRI data. Input data: 3D (whole-brain ROIs); outputs: group-level and subject-level networks.
Reference article: Kook et a. (2021). BVAR-Connect: A Variational Bayes Approach to Multi-Subject Vector Autoregressive Models for Inference on Brain Connectivity Networks. Neuroinformatics, 19, 39–56.
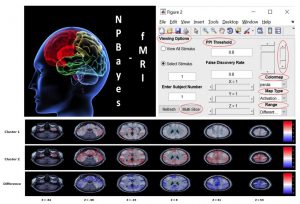
NPBayes-fMRI MATLAB GUI:User friendly software for non-parametric Bayesian spatio-temporal general linear models for task-based fMRI data. Input data: 2D (voxel-based) or 3D (whole-brain ROIs); pre-defined default setting for model parameters or customized choices; outputs: Subject-level activation maps, contrasts maps, and cluster-defined activation maps (for multiple subjects).
Reference article: Kook et al. (2019). NPBayes-fMRI: Nonparametric Bayesian General Linear Models for Single- and Multi-Subject fMRI Data. Stats in Biosciences, 11(1), 3-21.
Software at Bioconductor
 iClusterBayes – Bayesian clustering for multi-omics data. Available as part of iClusterPlus, developed as an enhanced version of the iCluster program of Shen, Olshen and Ladanyi (2009). The method employs latent variables for joint modeling of multiple omics data to cluster patient samples based on common latent factors. It produces posterior probabilities for each genomic feature to drive the integrative clustering.
iClusterBayes – Bayesian clustering for multi-omics data. Available as part of iClusterPlus, developed as an enhanced version of the iCluster program of Shen, Olshen and Ladanyi (2009). The method employs latent variables for joint modeling of multiple omics data to cluster patient samples based on common latent factors. It produces posterior probabilities for each genomic feature to drive the integrative clustering.
Reference article: Mo, Q., Shen, R., Guo, C., Vannucci, M., Chan, K. and Hilsenbeck, S.G. (2018). A Fully Bayesian Latent Variable Model for Integrative Clustering Analysis of Multi-type Omics Data. Biostatistics, 19(1), 71-86.
R Packages in CRAN
- IBATCGH – Integrative Bayesian Analysis of Transcriptomic and CGH Data
- KSCONS for Predicting Protein Residue Contact (shinyapp also available)
- SCRSELECT – Bayesian Variable Selection for Semi-Competing Risks
Chemometrics dataset:
NIR Spectral Data on Biscuit Doughs (available as part of the R package ppls)
Software for Protein Structure Prediction




